Chromosomes show off their shapes
Chromosomes -- the 46 tightly-wrapped packages of genetic material in our cells -- are iconically depicted as X-shaped formations. However, those neat X's only appear when a cell is about to divide and the entire contents of its genome duplicated. Until now researchers have not been able to get a good picture of the way that our DNA -- some two meters of strands all told -- is neatly bundled into the nucleus while enabling day-to-day (non-dividing) gene activity.
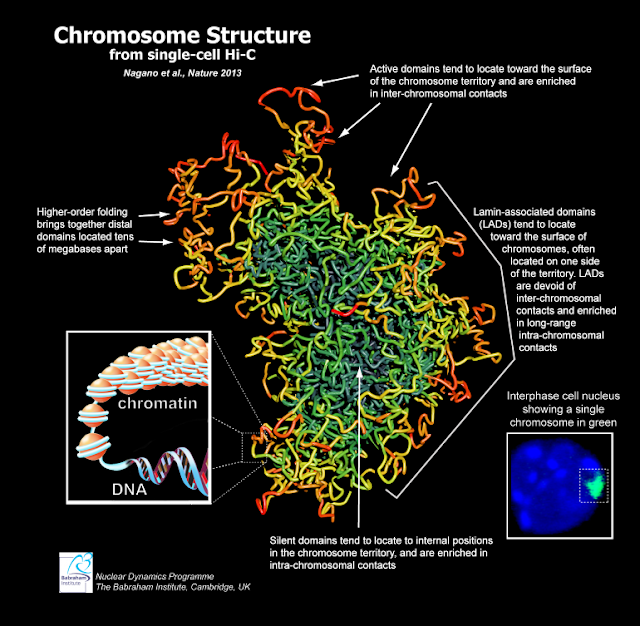
A combination of new techniques for sequencing DNA in individual chromosomes and analyzing data from thousands of measurements has given us a new picture of the 3-D structures of chromosomes. This method, the result of an international collaboration, which was recently reported in Nature, promises to help researchers understand the basic processes by which gene expression is regulated and genome stability maintained.
Prof. Amos Tanay of the Weizmann Institute's Computer Science and Applied Mathematics and the Biological Regulation Departments develops advanced computer algorithms for analyzing genomic datasets, which can run to billions of bits of information. He and his team, including PhD students Yaniv Lubling and Eitan Yaffe, joined forces with Dr. Peter Fraser of the Babraham Institute, UK, in an attempt to resolve chromosomal architectures at an unprecedentedly high resolution. Rather than the traditional microscopy techniques, they harnessed the power of modern high-throughput DNA sequencing. Fraser and his team developed a sophisticated sequencing method for taking thousands of measurements of the contacts between genes inside single cells.
While these techniques vastly improve upon approaches that average the conformations of millions of chromosomes, the data generated from just the few trillionths of a gram of DNA present within a single cell can only be interpreted by advanced statistical methods. Tanay and his team performed the complex computer analysis that turned millions of DNA sequences into reliable maps describing contacts between genes along individual chromosomes. Given these maps, the team, in collaboration with Dr. Ernest Laue of Cambridge University, UK, was able to produce 3-D models of individual chromosome structures.
Interestingly, the new high resolution depictions of chromosomal architecture indicate that the structure of the same DNA molecule can vary markedly between different cells. At the same time, the results point to some basic principles that underlie the genes' organization. Their arrangement appears to be modular and based on the functions of the thousands of genes embedded within each chromosome. The data suggests that chromosomes expose the more active genes at their boundaries, possibly allowing these genes better access to the cellular machinery that regulates them.
Besides giving us a unique, surprising view of the structure of the chromosomes in our cells, the researchers believe that their method will present genetics research with a powerful new tool. For example, it may help uncover the variations in genetic activity between different types of cells, or promote understanding of the mechanisms determining gene activity or quiescence in various normal or disease conditions. The rapidly increasing power of massive DNA sequencing promises to make studies such as this even more powerful in the near future.
Source: Weizmann Institute of Science [November 25, 2013]
 |
| The X-shape, often used to describe chromosomes, is only a snapshot of their complexity [Credit: istock] |
Prof. Amos Tanay of the Weizmann Institute's Computer Science and Applied Mathematics and the Biological Regulation Departments develops advanced computer algorithms for analyzing genomic datasets, which can run to billions of bits of information. He and his team, including PhD students Yaniv Lubling and Eitan Yaffe, joined forces with Dr. Peter Fraser of the Babraham Institute, UK, in an attempt to resolve chromosomal architectures at an unprecedentedly high resolution. Rather than the traditional microscopy techniques, they harnessed the power of modern high-throughput DNA sequencing. Fraser and his team developed a sophisticated sequencing method for taking thousands of measurements of the contacts between genes inside single cells.
While these techniques vastly improve upon approaches that average the conformations of millions of chromosomes, the data generated from just the few trillionths of a gram of DNA present within a single cell can only be interpreted by advanced statistical methods. Tanay and his team performed the complex computer analysis that turned millions of DNA sequences into reliable maps describing contacts between genes along individual chromosomes. Given these maps, the team, in collaboration with Dr. Ernest Laue of Cambridge University, UK, was able to produce 3-D models of individual chromosome structures.
Interestingly, the new high resolution depictions of chromosomal architecture indicate that the structure of the same DNA molecule can vary markedly between different cells. At the same time, the results point to some basic principles that underlie the genes' organization. Their arrangement appears to be modular and based on the functions of the thousands of genes embedded within each chromosome. The data suggests that chromosomes expose the more active genes at their boundaries, possibly allowing these genes better access to the cellular machinery that regulates them.
Besides giving us a unique, surprising view of the structure of the chromosomes in our cells, the researchers believe that their method will present genetics research with a powerful new tool. For example, it may help uncover the variations in genetic activity between different types of cells, or promote understanding of the mechanisms determining gene activity or quiescence in various normal or disease conditions. The rapidly increasing power of massive DNA sequencing promises to make studies such as this even more powerful in the near future.
Source: Weizmann Institute of Science [November 25, 2013]